| Overview | Cell Structures | Cell Migration | Cell Division |
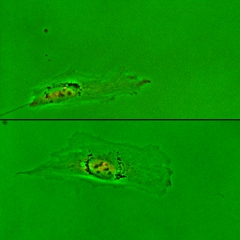
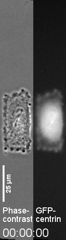
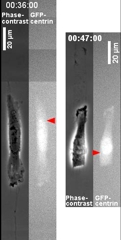
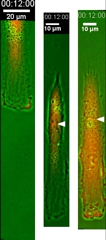
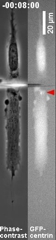
Control of the Location of the Rear Rather than Front by Centrosome |
Zhang et al., Mol. Biol. Cell. 28:3240-3251 (2018) |
Centrosome is often located in front of the nucleus relative to the direction of migration, which led to the conventional view that centrosome may dictate the position of the front. However there are many exceptions to this observation. We have proposed that microtubules may be responsible for the global distribution of inhibitory signals, away from the front to create a tail |